Three Way Chart HOWTO
Kenith Grey
2018-12-04
Required Libraries
require(ggplot2)
require(ggQC)
require(gridExtra)set.seed(5555)
Process1 <- data.frame(ID = 1:100, process = rep(1,100), value = rnorm(100,0,1), repitition=1:20 )
Process2 <- data.frame(ID = 101:200, process = rep(2,100), value = rnorm(100,5,.5), repitition=1:10 )
all_processes <- rbind(Process1, Process2)Simple Three Way Chart
ggX <- ggplot(data = Process1, aes(x=repitition, y=value)) +
stat_QC(method="xBar.sBar", auto.label = T, label.digits = 3)
ggmR <- ggplot(data = Process1, aes(x=repitition,y=value)) +
stat_QC(method = "mR", auto.label = T, label.digits = 2) +
ylab("mR")
ggRbar <- ggplot(data = Process1, aes(x=repitition,y=value)) +
stat_QC(method="rBar", auto.label = T, label.digits = 3) +
ylab("Rbar")
grid.arrange(ggX, ggmR, ggRbar, nrow=3)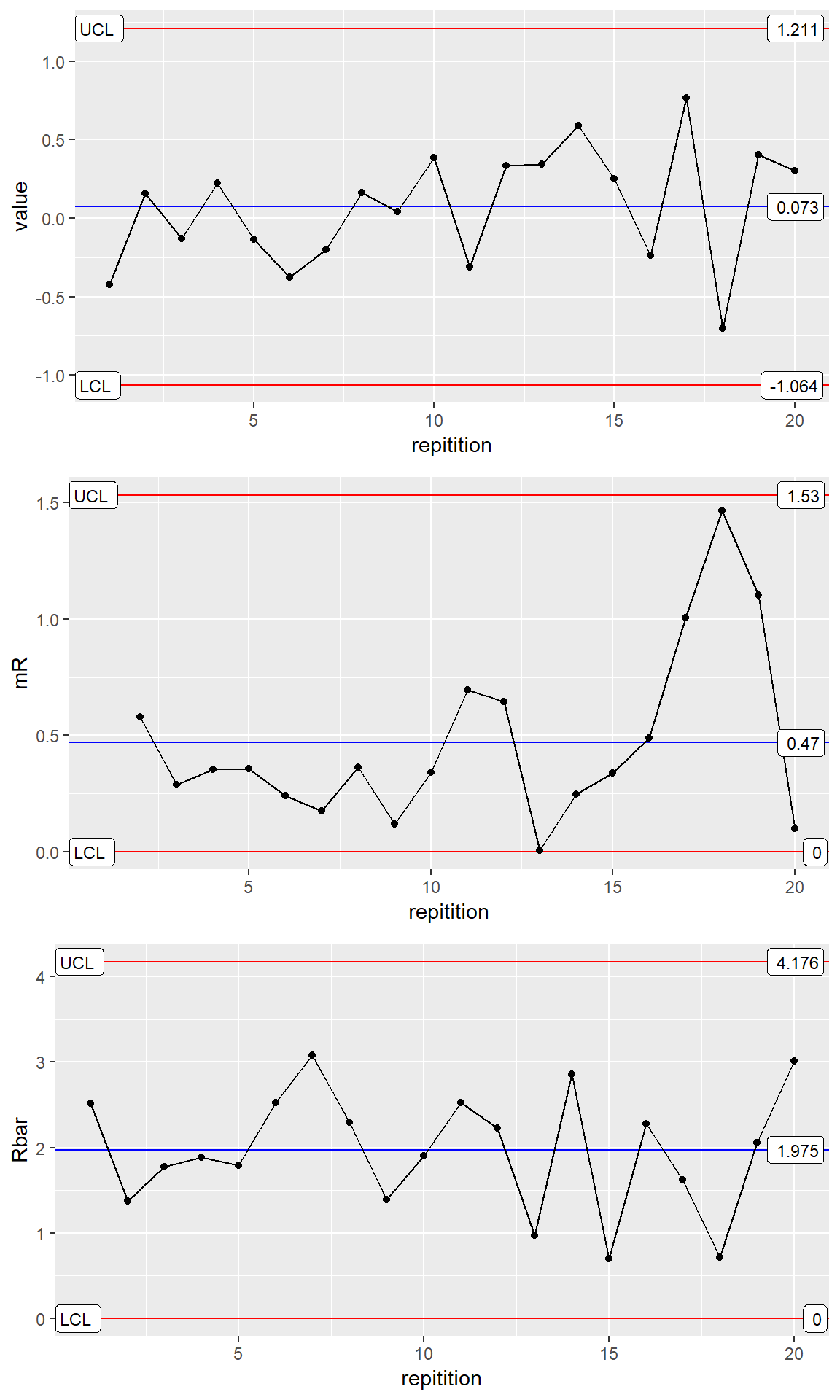
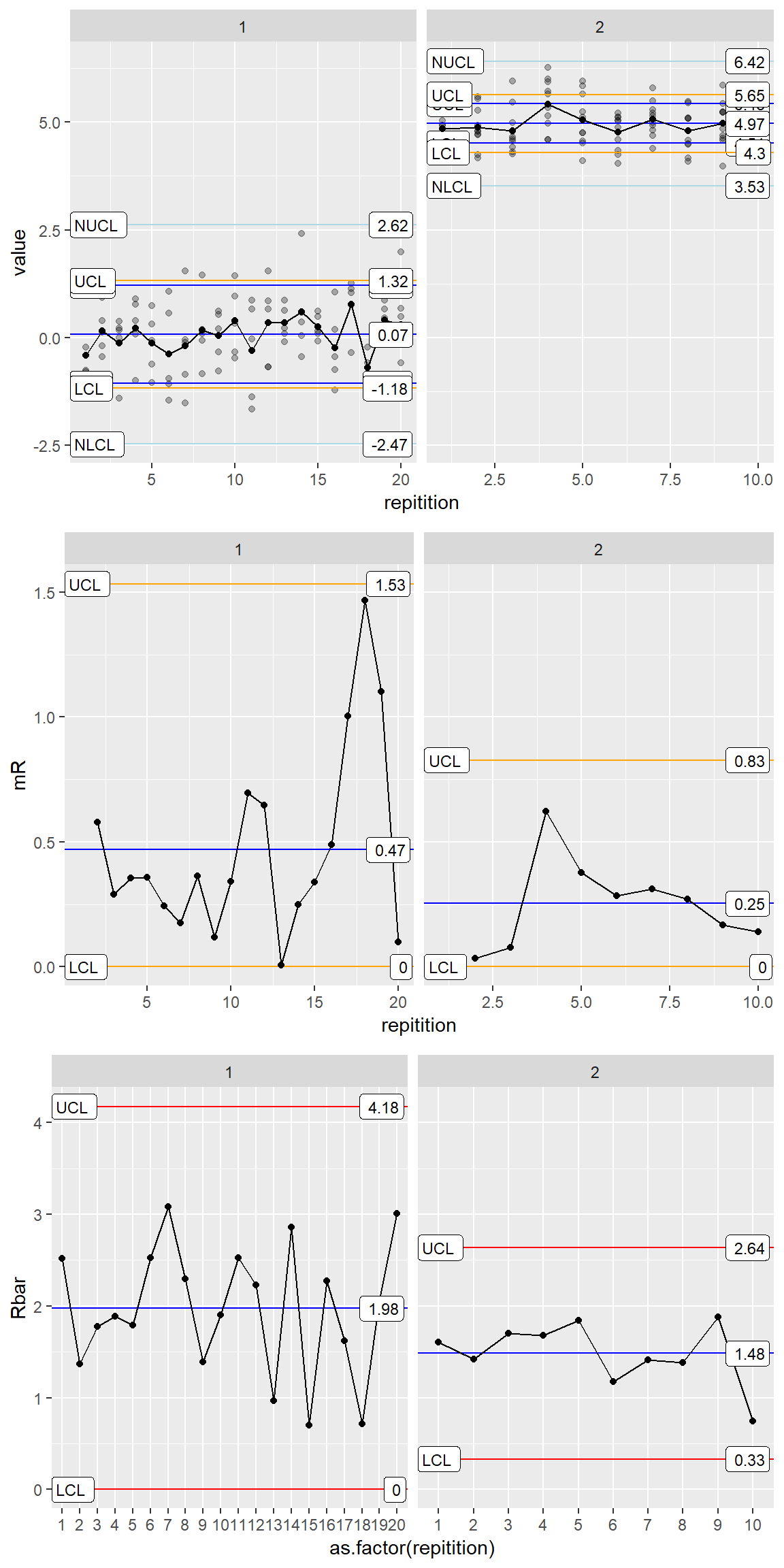
Complex Three Way Chart
ggX <- ggplot(data = all_processes, aes(x=repitition, y=value)) +
geom_point(alpha=.3) +
stat_QC(method="xBar.rBar", color.qc_limits = "blue", auto.label = T, label.digits = 2) +
stat_QC(method="XmR", color.qc_limits = "orange", auto.label = T, label.digits = 2) +
stat_QC(method="xBar.rBar", n = 1, color.qc_limits = "lightblue",
auto.label = T, limit.txt.label = c("NLCL", "NUCL"), label.digits = 2 ) +
facet_grid(.~process, scales = "free_x")
ggmR <- ggplot(data = all_processes, aes(x=repitition,y=value)) +
stat_QC(method="mR", color.qc_limits = "orange", auto.label = T, label.digits = 2) +
ylab("mR") +
facet_grid(.~process, scales = "free_x")
ggRbar <- ggplot(data = all_processes, aes(x=as.factor(repitition),y=value, group=process)) +
stat_QC(method="rBar", auto.label = T, label.digits = 2) +
ylab("Rbar") +
facet_grid(.~process, scales = "free_x")
grid.arrange(ggX, ggmR, ggRbar, nrow=3)